Welcome to BacArena!
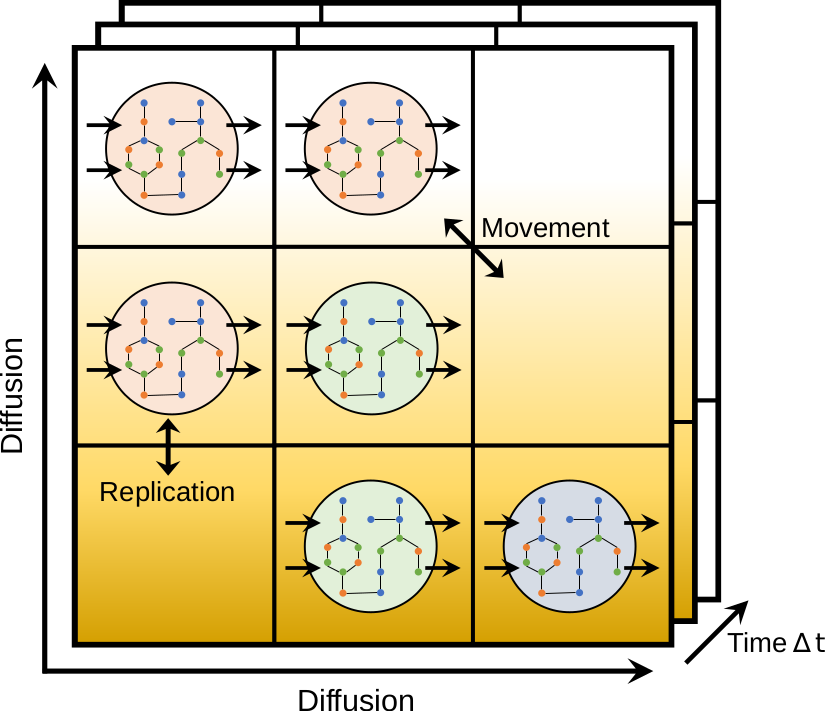
BacArena is an open source software for simulating cellular communities. It combines agent-based modeling, flux balance analysis, and statistical analysis. BacArena is implemented in R and available on CRAN.
Get the latest news and updates:
GitHub
Check out the BacArena publication:
Eugen Bauer*, Johannes Zimmermann*, Federico Baldini, Ines Thiele, Christoph Kaleta
BacArena: Individual-based metabolic modeling of heterogeneous microbes in complex communities PLOS Computational Biology (2017) 13,5 doi:10.1371/journal.pcbi.1005544
Installation and first steps
BacArena is written in R language and can be installed easily via The Comprehensive R Archive Network (CRAN):
install.packages('BacArena')
library('BacArena')
openArena()
Installing the development version
The CRAN version of BacArena is updated less frequently.
If you want to use the latest features and bugfixes, then try out the developtment version:
install.packages('devtools')
install.github("euba/bacarena")
library('BacArena')
Import matlab models
matlab cobra model files can be loaded into BacArena:
mod <- readMATmod("model.mat")
Check out the tutorial for details.
Problems with SBML
Currently, sybilSBML is not available on CRAN.
You can install it manually from a linux terminal by:
wget https://www.cs.hhu.de/fileadmin/redaktion/Oeffentliche_Medien/Fakultaeten/Mathematisch-Naturwissenschaftliche_Fakultaet/Informatik/Bioinformatik/sybilSBML_3.0.5.tar.gz
R CMD INSTALL sybilSBML_3.0.5.tar.gz
Getting help
Check out our Tutorial and Manual
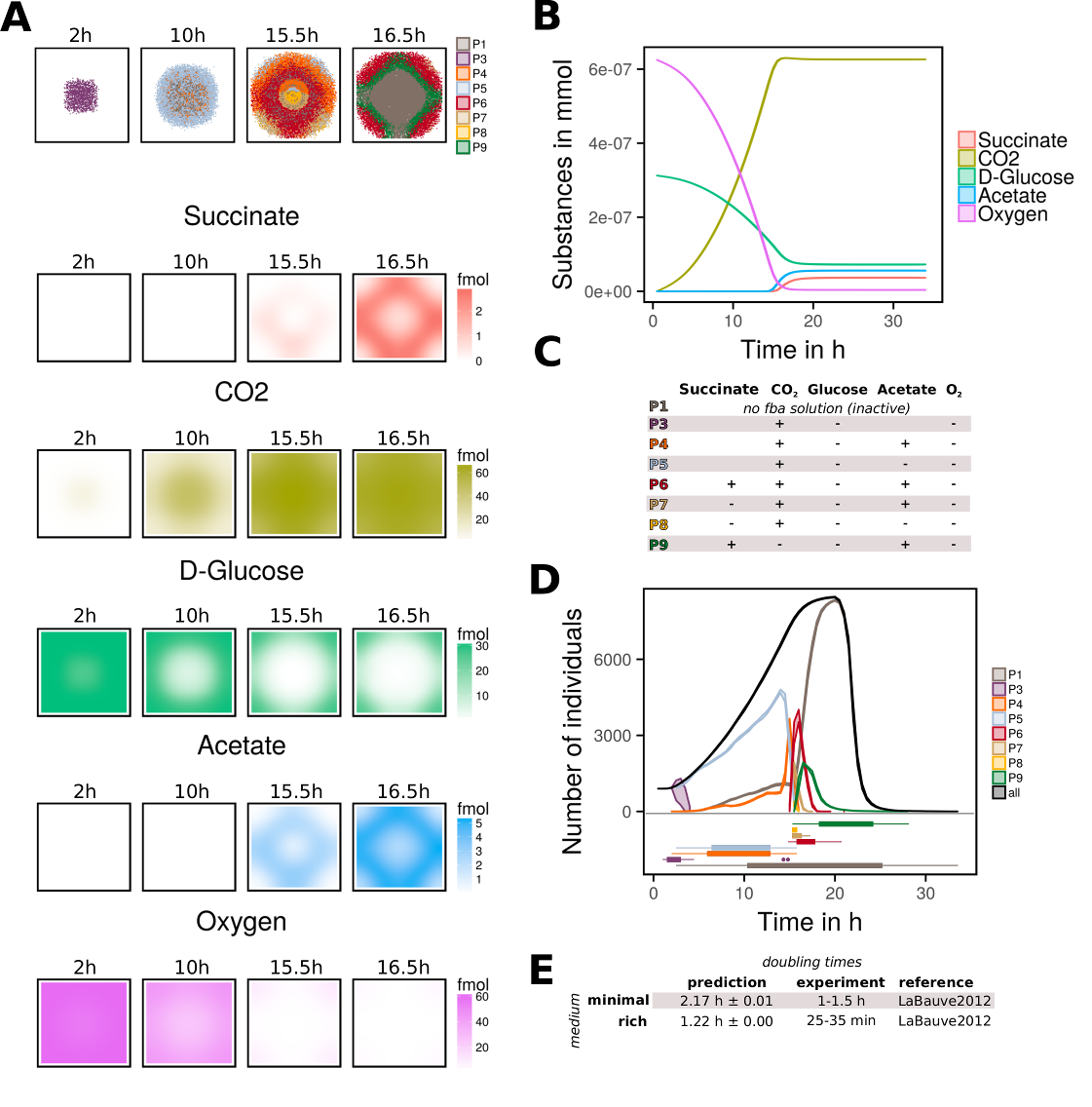
- Doing a simple simulation using the E. coli model (iJO1366)
- Working with flux distributions and reaction activities
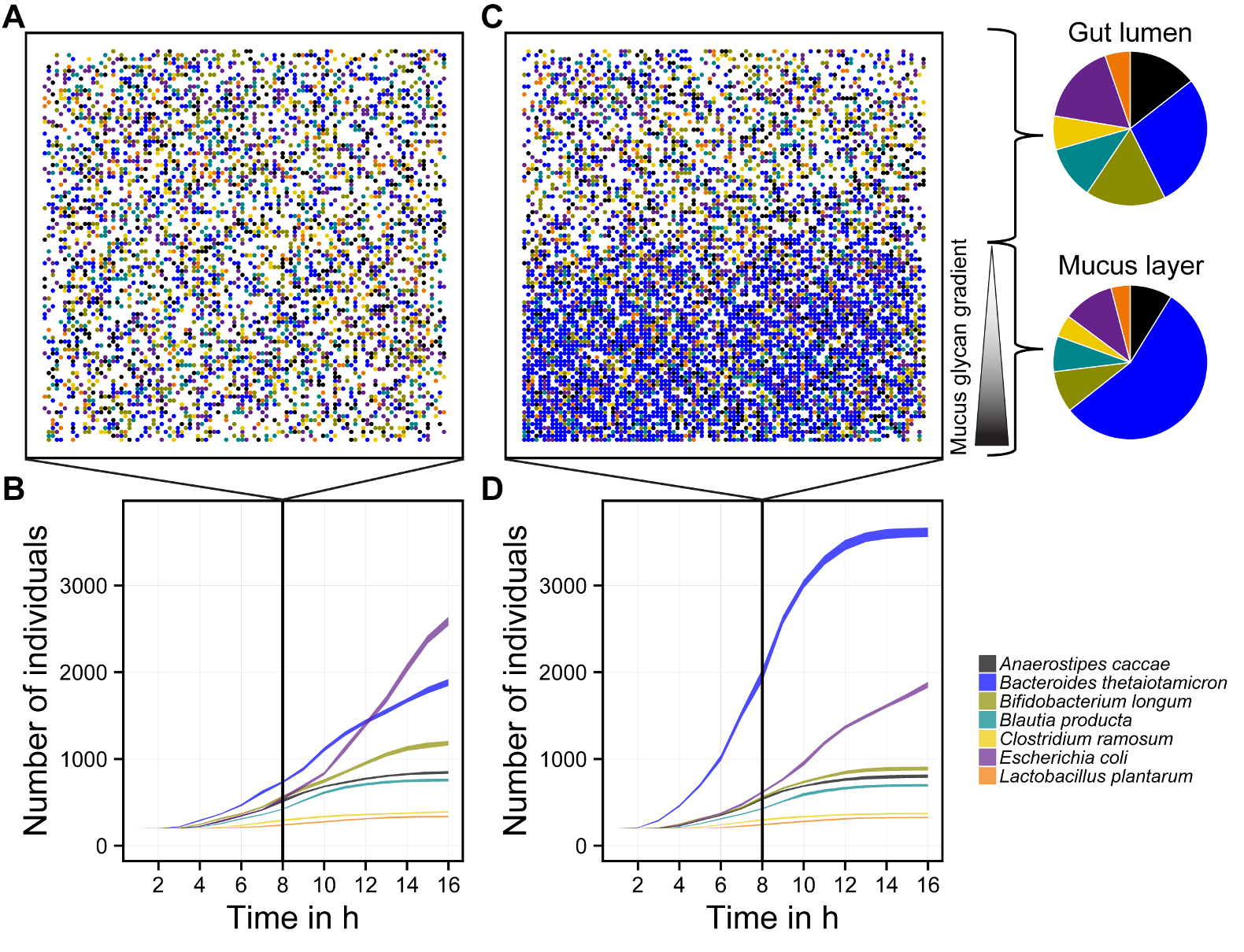
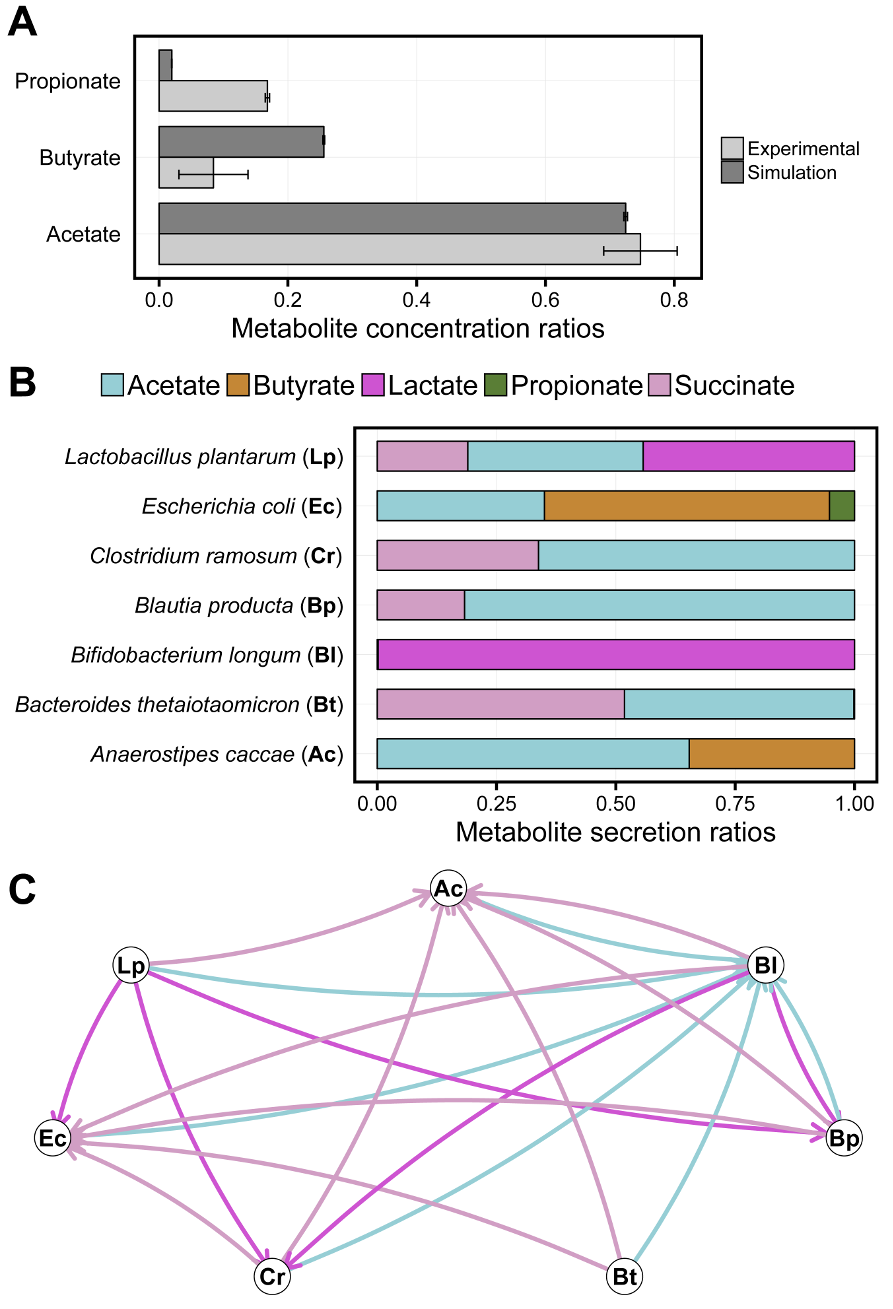
- Analyzing bacterial interaction and cross-feeding patterns
- Simple simulation without spatial component and disabled growth model
- model environmental impact by switching from anaerobic to aerobic conditions after some time
- how to import matlab models
Pictures